Thank you for the many requests to provide some extra info on how best to get `finalfit` results out of RStudio, and particularly into Microsoft Word.
Here is how.
Make sure you are on the most up-to-date version of `finalfit`.
devtools::install_github("ewenharrison/finalfit")
What follows is for demonstration purposes and is not meant to illustrate model building.
Does a tumour characteristic (differentiation) predict 5-year survival?
Demographics table
First explore variable of interest (exposure) by making it the dependent.
library(finalfit)
library(dplyr)
dependent = "differ.factor"
# Specify explanatory variables of interest
explanatory = c("age", "sex.factor",
"extent.factor", "obstruct.factor",
"nodes")
Note this useful alternative way of specifying explanatory variable lists:
colon_s %>% select(age, sex.factor, extent.factor, obstruct.factor, nodes) %>% names() -> explanatory
Look at associations between our exposure and other explanatory variables. Include missing data.
colon_s %>% summary_factorlist(dependent, explanatory, p=TRUE, na_include=TRUE)
label levels Well Moderate Poor p
Age (years) Mean (SD) 60.2 (12.8) 59.9 (11.7) 59 (12.8) 0.788
Sex Female 51 (11.6) 314 (71.7) 73 (16.7) 0.400
Male 42 (9.0) 349 (74.6) 77 (16.5)
Extent of spread Submucosa 5 (25.0) 12 (60.0) 3 (15.0) 0.081
Muscle 12 (11.8) 78 (76.5) 12 (11.8)
Serosa 76 (10.2) 542 (72.8) 127 (17.0)
Adjacent structures 0 (0.0) 31 (79.5) 8 (20.5)
Obstruction No 69 (9.7) 531 (74.4) 114 (16.0) 0.110
Yes 19 (11.0) 122 (70.9) 31 (18.0)
Missing 5 (25.0) 10 (50.0) 5 (25.0)
nodes Mean (SD) 2.7 (2.2) 3.6 (3.4) 4.7 (4.4) <0.001
Warning messages:
1: In chisq.test(tab, correct = FALSE) :
Chi-squared approximation may be incorrect
2: In chisq.test(tab, correct = FALSE) :
Chi-squared approximation may be incorrect
Note missing data in `obstruct.factor`. We will drop this variable for now (again, this is for demonstration only). Also see that `nodes` has not been labelled.
There are small numbers in some variables generating chisq.test warnings (predicted less than 5 in any cell). Generate final table.
Hmisc::label(colon_s$nodes) = "Lymph nodes involved"
explanatory = c("age", "sex.factor",
"extent.factor", "nodes")
colon_s %>%
summary_factorlist(dependent, explanatory,
p=TRUE, na_include=TRUE,
add_dependent_label=TRUE) -> table1
table1
Dependent: Differentiation Well Moderate Poor p
Age (years) Mean (SD) 60.2 (12.8) 59.9 (11.7) 59 (12.8) 0.788
Sex Female 51 (11.6) 314 (71.7) 73 (16.7) 0.400
Male 42 (9.0) 349 (74.6) 77 (16.5)
Extent of spread Submucosa 5 (25.0) 12 (60.0) 3 (15.0) 0.081
Muscle 12 (11.8) 78 (76.5) 12 (11.8)
Serosa 76 (10.2) 542 (72.8) 127 (17.0)
Adjacent structures 0 (0.0) 31 (79.5) 8 (20.5)
Lymph nodes involved Mean (SD) 2.7 (2.2) 3.6 (3.4) 4.7 (4.4) <0.001
Logistic regression table
Now examine explanatory variables against outcome. Check plot runs ok.
explanatory = c("age", "sex.factor",
"extent.factor", "nodes",
"differ.factor")
dependent = "mort_5yr"
colon_s %>%
finalfit(dependent, explanatory,
dependent_label_prefix = "") -> table2
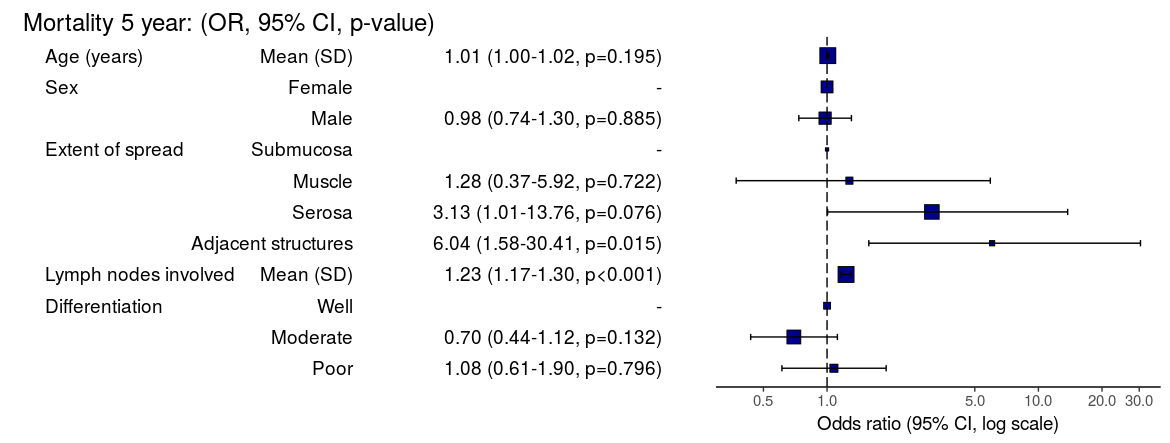
Mortality 5 year Alive Died OR (univariable) OR (multivariable)
Age (years) Mean (SD) 59.8 (11.4) 59.9 (12.5) 1.00 (0.99-1.01, p=0.986) 1.01 (1.00-1.02, p=0.195)
Sex Female 243 (47.6) 194 (48.0) - -
Male 268 (52.4) 210 (52.0) 0.98 (0.76-1.27, p=0.889) 0.98 (0.74-1.30, p=0.885)
Extent of spread Submucosa 16 (3.1) 4 (1.0) - -
Muscle 78 (15.3) 25 (6.2) 1.28 (0.42-4.79, p=0.681) 1.28 (0.37-5.92, p=0.722)
Serosa 401 (78.5) 349 (86.4) 3.48 (1.26-12.24, p=0.027) 3.13 (1.01-13.76, p=0.076)
Adjacent structures 16 (3.1) 26 (6.4) 6.50 (1.98-25.93, p=0.004) 6.04 (1.58-30.41, p=0.015)
Lymph nodes involved Mean (SD) 2.7 (2.4) 4.9 (4.4) 1.24 (1.18-1.30, p<0.001) 1.23 (1.17-1.30, p<0.001)
Differentiation Well 52 (10.5) 40 (10.1) - -
Moderate 382 (76.9) 269 (68.1) 0.92 (0.59-1.43, p=0.694) 0.70 (0.44-1.12, p=0.132)
Poor 63 (12.7) 86 (21.8) 1.77 (1.05-3.01, p=0.032) 1.08 (0.61-1.90, p=0.796)
Odds ratio plot
colon_s %>% or_plot(dependent, explanatory, breaks = c(0.5, 1, 5, 10, 20, 30))
To MS Word via knitr/R Markdown
Important. In most R Markdown set-ups, environment objects require to be saved and loaded to R Markdown document.
# Save objects for knitr/markdown save(table1, table2, dependent, explanatory, file = "out.rda")
We use RStudio Server Pro set-up on Ubuntu. But these instructions should work fine for most/all RStudio/Markdown default set-ups.
In RStudio, select `File > New File > R Markdown`.
A useful template file is produced by default. Try hitting `knit to Word` on the `knitr` button at the top of the `.Rmd` script window.
Now paste this into the file:
---
title: "Example knitr/R Markdown document"
author: "Ewen Harrison"
date: "22/5/2018"
output:
word_document: default
---
```{r setup, include=FALSE}
# Load data into global environment.
library(finalfit)
library(dplyr)
library(knitr)
load("out.rda")
```
## Table 1 - Demographics
```{r table1, echo = FALSE, results='asis'}
kable(table1, row.names=FALSE, align=c("l", "l", "r", "r", "r", "r"))
```
## Table 2 - Association between tumour factors and 5 year mortality
```{r table2, echo = FALSE, results='asis'}
kable(table2, row.names=FALSE, align=c("l", "l", "r", "r", "r", "r"))
```
## Figure 1 - Association between tumour factors and 5 year mortality
```{r figure1, echo = FALSE}
colon_s %>%
or_plot(dependent, explanatory)
```
Create Word template file
Now, edit the Word template. Click on a table. The `style` should be `compact`. Right click > `Modify... > font size = 9`. Alter heading and text styles in the same way as desired. Save this as `template.docx`. Upload to your project folder. Add this reference to the `.Rmd` YAML heading, as below. Make sure you get the space correct.
The plot also doesn't look quite right and it prints with warning messages. Experiment with `fig.width` to get it looking right.
Now paste this into your `.Rmd` file and run:
---
title: "Example knitr/R Markdown document"
author: "Ewen Harrison"
date: "21/5/2018"
output:
word_document:
reference_docx: template.docx
---
```{r setup, include=FALSE}
# Load data into global environment.
library(finalfit)
library(dplyr)
library(knitr)
load("out.rda")
```
## Table 1 - Demographics
```{r table1, echo = FALSE, results='asis'}
kable(table1, row.names=FALSE, align=c("l", "l", "r", "r", "r", "r"))
```
## Table 2 - Association between tumour factors and 5 year mortality
```{r table2, echo = FALSE, results='asis'}
kable(table2, row.names=FALSE, align=c("l", "l", "r", "r", "r", "r"))
```
## Figure 1 - Association between tumour factors and 5 year mortality
```{r figure1, echo = FALSE, warning=FALSE, message=FALSE, fig.width=10}
colon_s %>%
or_plot(dependent, explanatory)
```
This is now looking good for me, and further tweaks can be made.
To PDF via knitr/R Markdown
Default settings for PDF:
---
title: "Example knitr/R Markdown document"
author: "Ewen Harrison"
date: "21/5/2018"
output:
pdf_document: default
---
```{r setup, include=FALSE}
# Load data into global environment.
library(finalfit)
library(dplyr)
library(knitr)
load("out.rda")
```
## Table 1 - Demographics
```{r table1, echo = FALSE, results='asis'}
kable(table1, row.names=FALSE, align=c("l", "l", "r", "r", "r", "r"))
```
## Table 2 - Association between tumour factors and 5 year mortality
```{r table2, echo = FALSE, results='asis'}
kable(table2, row.names=FALSE, align=c("l", "l", "r", "r", "r", "r"))
```
## Figure 1 - Association between tumour factors and 5 year mortality
```{r figure1, echo = FALSE}
colon_s %>%
or_plot(dependent, explanatory)
```
Again, ok but not great.
[gview file="http://www.datasurg.net/wp-content/uploads/2018/05/example.pdf"]
We can fix the plot in exactly the same way. But the table is off the side of the page. For this we use the `kableExtra` package. Install this in the normal manner. You may also want to alter the margins of your page using `geometry` in the preamble.
---
title: "Example knitr/R Markdown document"
author: "Ewen Harrison"
date: "21/5/2018"
output:
pdf_document: default
geometry: margin=0.75in
---
```{r setup, include=FALSE}
# Load data into global environment.
library(finalfit)
library(dplyr)
library(knitr)
library(kableExtra)
load("out.rda")
```
## Table 1 - Demographics
```{r table1, echo = FALSE, results='asis'}
kable(table1, row.names=FALSE, align=c("l", "l", "r", "r", "r", "r"),
booktabs=TRUE)
```
## Table 2 - Association between tumour factors and 5 year mortality
```{r table2, echo = FALSE, results='asis'}
kable(table2, row.names=FALSE, align=c("l", "l", "r", "r", "r", "r"),
booktabs=TRUE) %>%
kable_styling(font_size=8)
```
## Figure 1 - Association between tumour factors and 5 year mortality
```{r figure1, echo = FALSE, warning=FALSE, message=FALSE, fig.width=10}
colon_s %>%
or_plot(dependent, explanatory)
```
This is now looking pretty good for me as well.
[gview file="http://www.datasurg.net/wp-content/uploads/2018/05/example2.pdf"]
There you have it. A pretty quick workflow to get final results into Word and a PDF.

Can you please specify the data source for this table? If its confidential, maybe just the first first few observations so I can make a sample data set and try out your code on my own. Thanks
The data is included in the package. It should all run as is. If not, add `data(colon_s)`.
Is there an easy way to reduce the text size in or_plot?
Hi,
Yes, make sure you are on the most up-to-date version. Any ggplot argument can be passed via `plot_opts`. The `table_text_size` etc. can be adjusted directly.
Ewen
Regarding:
> Any ggplot argument can be passed via plot_opts
I’m sorry, could you post an example of passing a ggplot argument via plot_opts to or_plot()?
I tried a few different things and was unable to alter the plot output — I assume it’s something very obvious that I could figure out by more trial-and-error, but at this point, I think it’d be quicker to just ask, and then others who find this page could also see an example.
There is an example on the `or_plot` help page.
“`
colon_s %>%
or_plot(dependent, explanatory,
table_text_size=4,
title_text_size=14,
plot_opts=list(xlab(“OR, 95% CI”),
theme(axis.title = element_text(size=12))))
“`
Thank you, but when I run that code, the x-label and text sizes don’t change.
I get the identical plot as with just colon_s %>% or_plot(dependent, explanatory)
Update (can’t seem to edit my reply): code to adjust text size / axis labels worked after updating to latest version with:
devtools::install_github(“ewenharrison/finalfit”)
Thanks very much! Very helpful package!
This package is AMAZING!!!
It will revolutionize my PhD.
Thank you for sharing
Hi,
Thanks for sharing this.
However, I noticed that the “Odds Ratio” output of the table is different from the values I obtained when I run logistic regression algorithm directly in R or Stata.
Can you kindly confirm if the “OR” in the output table is Odds Ratio? If not, how can I convert to Odds Ratio
Thanks. The table output is from logistic regression run directly in R, so it will be the same.
Please provide reproducible example if think it is not working properly.